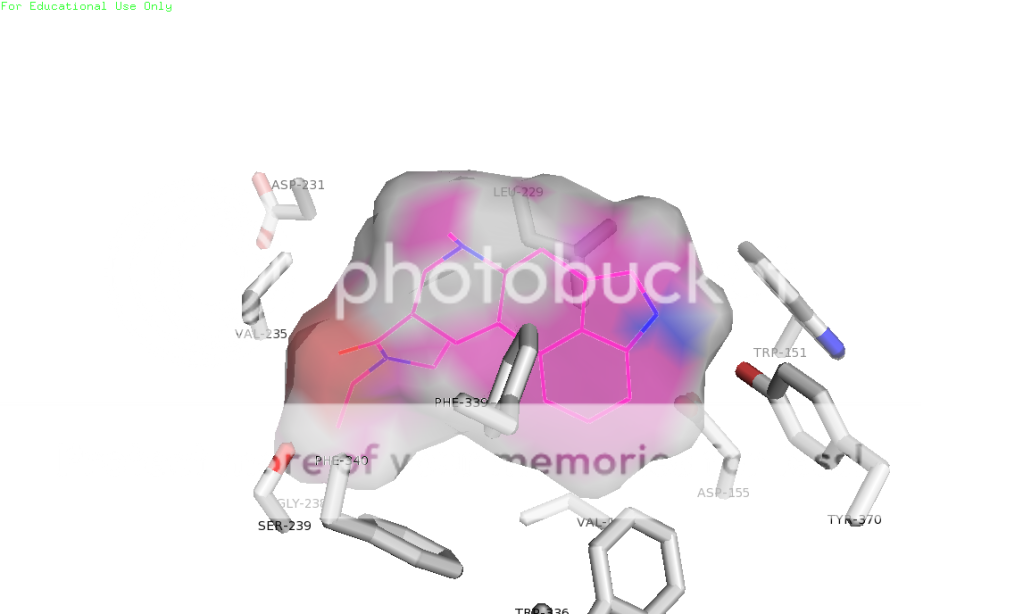
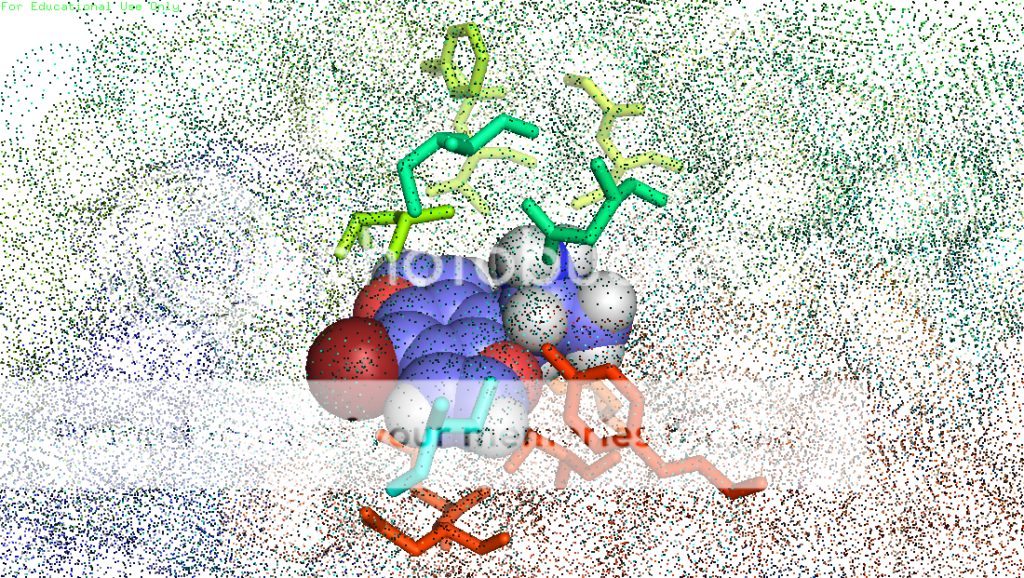
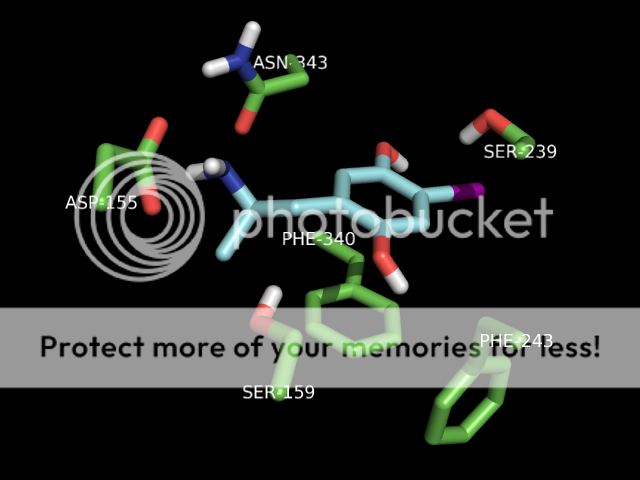
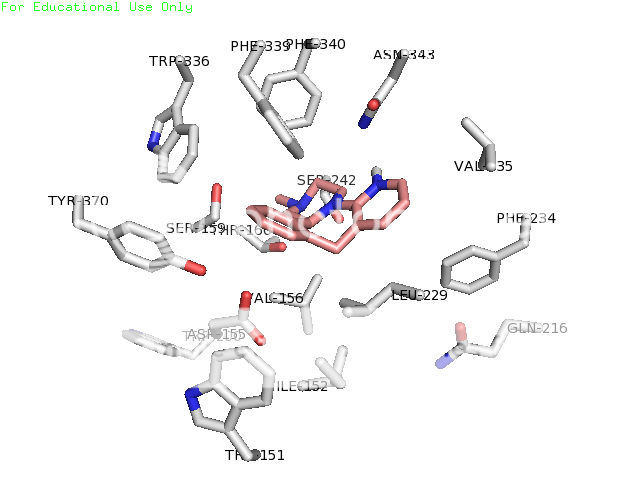
thanks skillet, i would be very grateful if you could slip me that pdb. maybe i could get that lsd pose right! also, when using autodock programs, you don't just get one pose usually. you get a few, mostly the same, maybe a slightly different conformer or something, but lsd is known to have more than one binding mode in 5ht2a (if i understand correctly). this is one of the reasons for it's uncommon potency. i don't remember how many i got for lsd, but i do know i chose the one to render in pymol based on it's similarity to the image in a 2008 paper so it would look good to the department bigwigs, who are not necessarily lsd chemistry experts, right? lemme dig thru and see if i have a pose more to your liking ; )
also, you're absolutely right about consistency in rendering style. some of those shots are from when i was first learning to do this, so i was experimenting. now i know how to get the poses neatly composed and residue labels, etc. Any other ideas about this are welcome as constructive criticism. and i've been lurking for a WHILE, have seen some of your posts. you're one of the good ones.
when i put up ibogaine or mirtazapine (hopefully soon, but it's monday) those will be all new docking/rendering so check the technique and feedback is appreciated. all of you.
thanks,
nirvus