mitogen
Bluelighter
- Joined
- Nov 8, 2004
- Messages
- 127
I've just spent the last two hours working out how to write scripts in PyMOL (its a Python based macromolecular structure visualisation program)
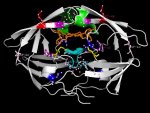
Attached is my masterpiece, the HIV Protease structure with annotated amino acids.
It's color coded: purple, blue, yellow and red amino acids are where protease inhibitor induced mutational hotspots occur. From a list of current antiHIV1_protease drugs I compiled what drugs induce what mutations and how common these mutations are among drugs. purple = 1, blue =2, yellow = 3, red =4.
Additionally, the active site (Asp25, Thr26, Gly27) is coloured cyan
The inhibitor molecule that was complexed with HIV1_PR (ritonavir) is colored orange
Two Ile residues important for docking are colored green.
The majority (~9/15) of HIV viral proteins are created as polyproteins. Imagine a plastic model airplane: when you buy it all the components are in a kind of plastic rack and you have to cut them out with a scalpel and smooth off the edges. Think of HIV_PR as that scalpel: first it cuts itself out of the rack, then it cuts the other proteins out. This process is the maturation of the virus: once the proteins are cut out of the rack they self-assemble. Simulatneous to maturation, the virion, which is now located on the cell surface, buds off the surface to go find another cell to infect.
I know this is slightly OT, as it is regarding HIV drugs, but this is the kind of work people do when they have a structure and they want to find out how to manipulate it with small molecules.
Currently it's a bitch to get high resolution crystal structures of membrane proteins (ie most receptors,) but one day we'll get the techniques sorted and then we'll be able to do this kind of work with membrane proteins, and continue this work with psychoactive drugs.
Attached is my masterpiece, the HIV Protease structure with annotated amino acids.
It's color coded: purple, blue, yellow and red amino acids are where protease inhibitor induced mutational hotspots occur. From a list of current antiHIV1_protease drugs I compiled what drugs induce what mutations and how common these mutations are among drugs. purple = 1, blue =2, yellow = 3, red =4.
Additionally, the active site (Asp25, Thr26, Gly27) is coloured cyan
The inhibitor molecule that was complexed with HIV1_PR (ritonavir) is colored orange
Two Ile residues important for docking are colored green.
The majority (~9/15) of HIV viral proteins are created as polyproteins. Imagine a plastic model airplane: when you buy it all the components are in a kind of plastic rack and you have to cut them out with a scalpel and smooth off the edges. Think of HIV_PR as that scalpel: first it cuts itself out of the rack, then it cuts the other proteins out. This process is the maturation of the virus: once the proteins are cut out of the rack they self-assemble. Simulatneous to maturation, the virion, which is now located on the cell surface, buds off the surface to go find another cell to infect.
I know this is slightly OT, as it is regarding HIV drugs, but this is the kind of work people do when they have a structure and they want to find out how to manipulate it with small molecules.
Currently it's a bitch to get high resolution crystal structures of membrane proteins (ie most receptors,) but one day we'll get the techniques sorted and then we'll be able to do this kind of work with membrane proteins, and continue this work with psychoactive drugs.